1. Functional genes
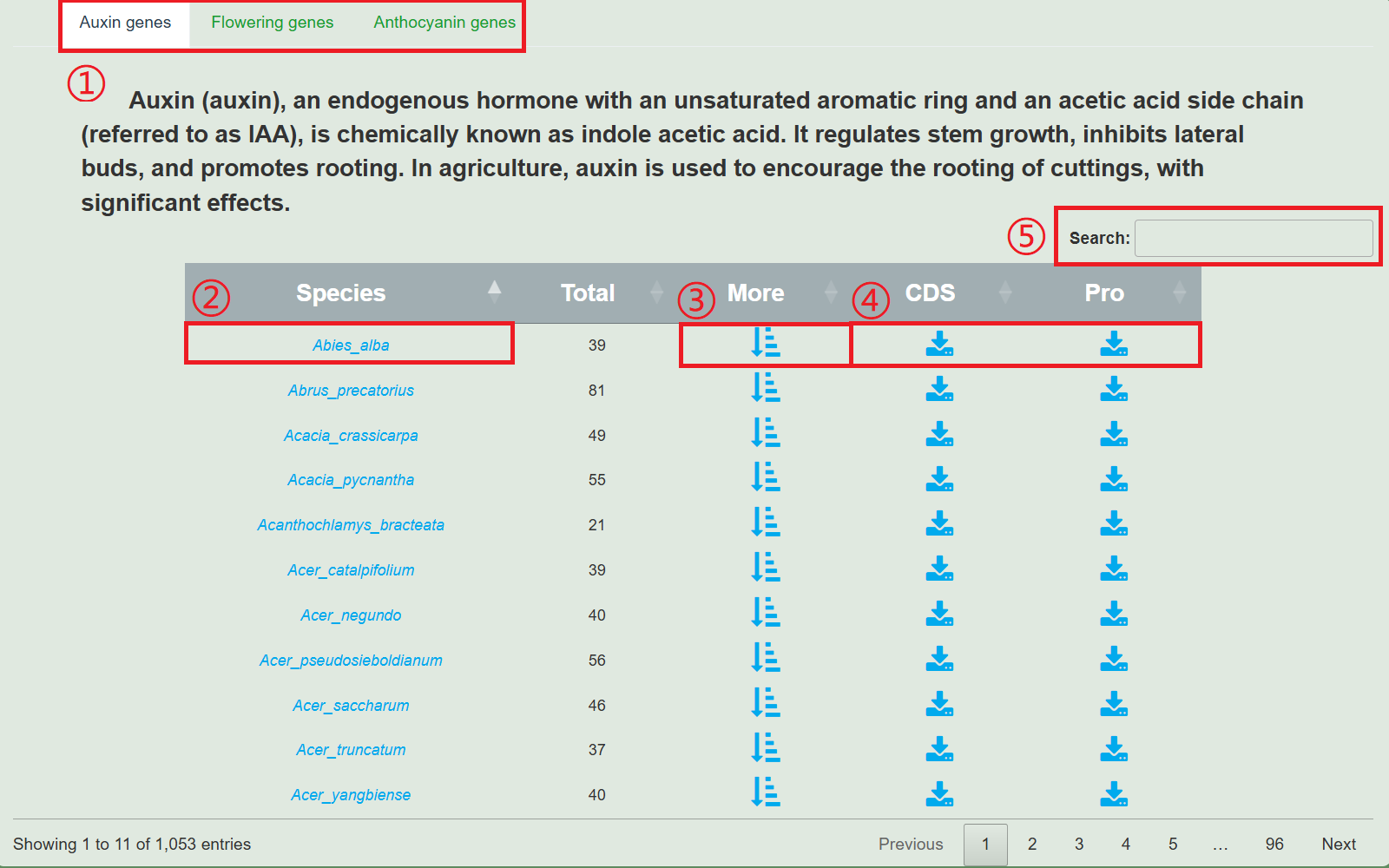
1.Switch between three gene types.
2.Clicking on the species name will redirect you to the species display page.
3.Click "More" to view the functional genes of this species, pop up the following image.
4.Download the CDS or protein sequence of the functional gene of this species.
5.Search for species.
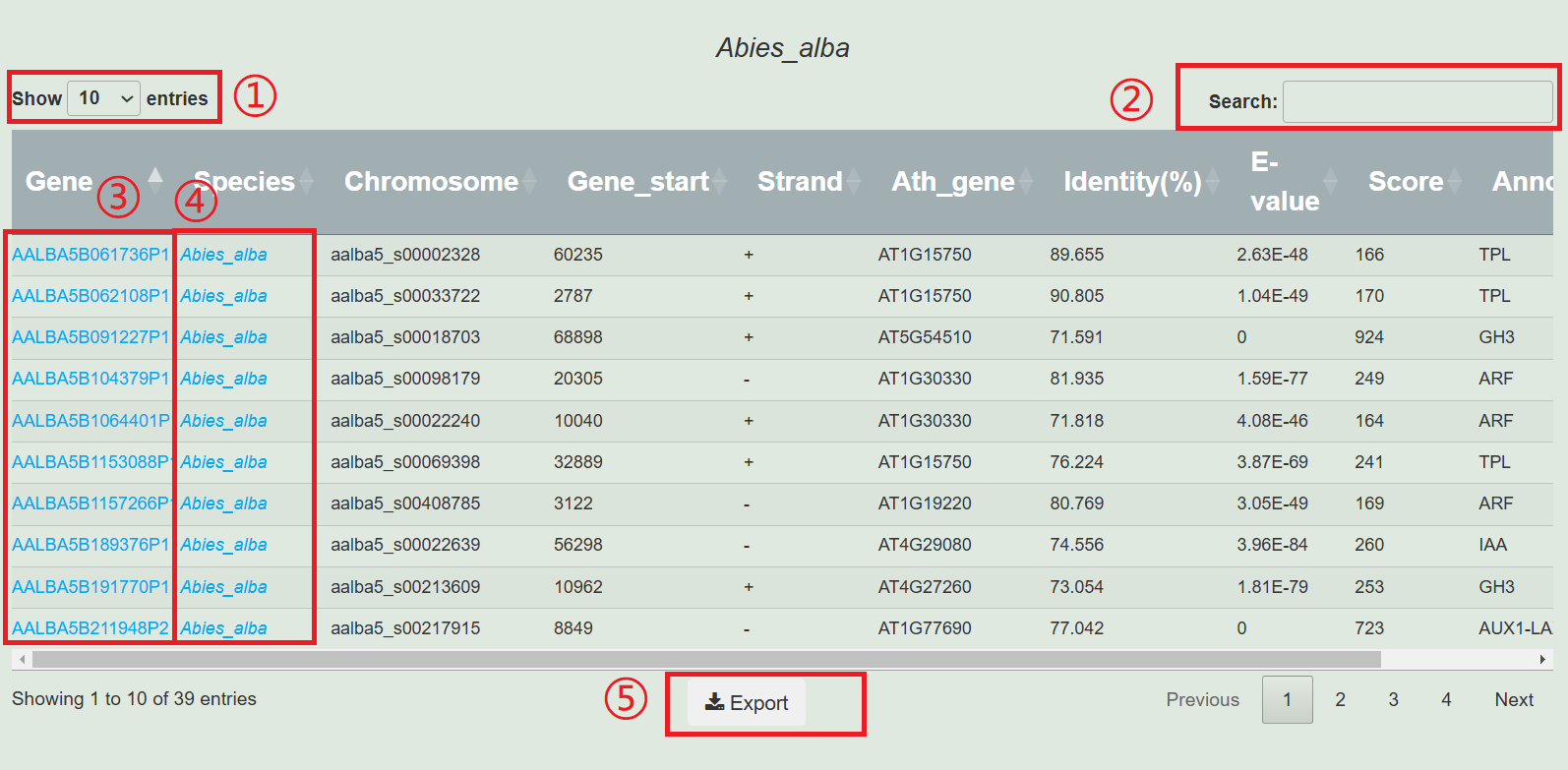
1.Select the number of displays.
2.Input genes, chromosomes, and annotations of interest for screening.
3.Click on the gene to jump to the gene display page.
4.Clicking on the species name will redirect you to the species display page.
5.Download all data from the table.
2. Transcription factors
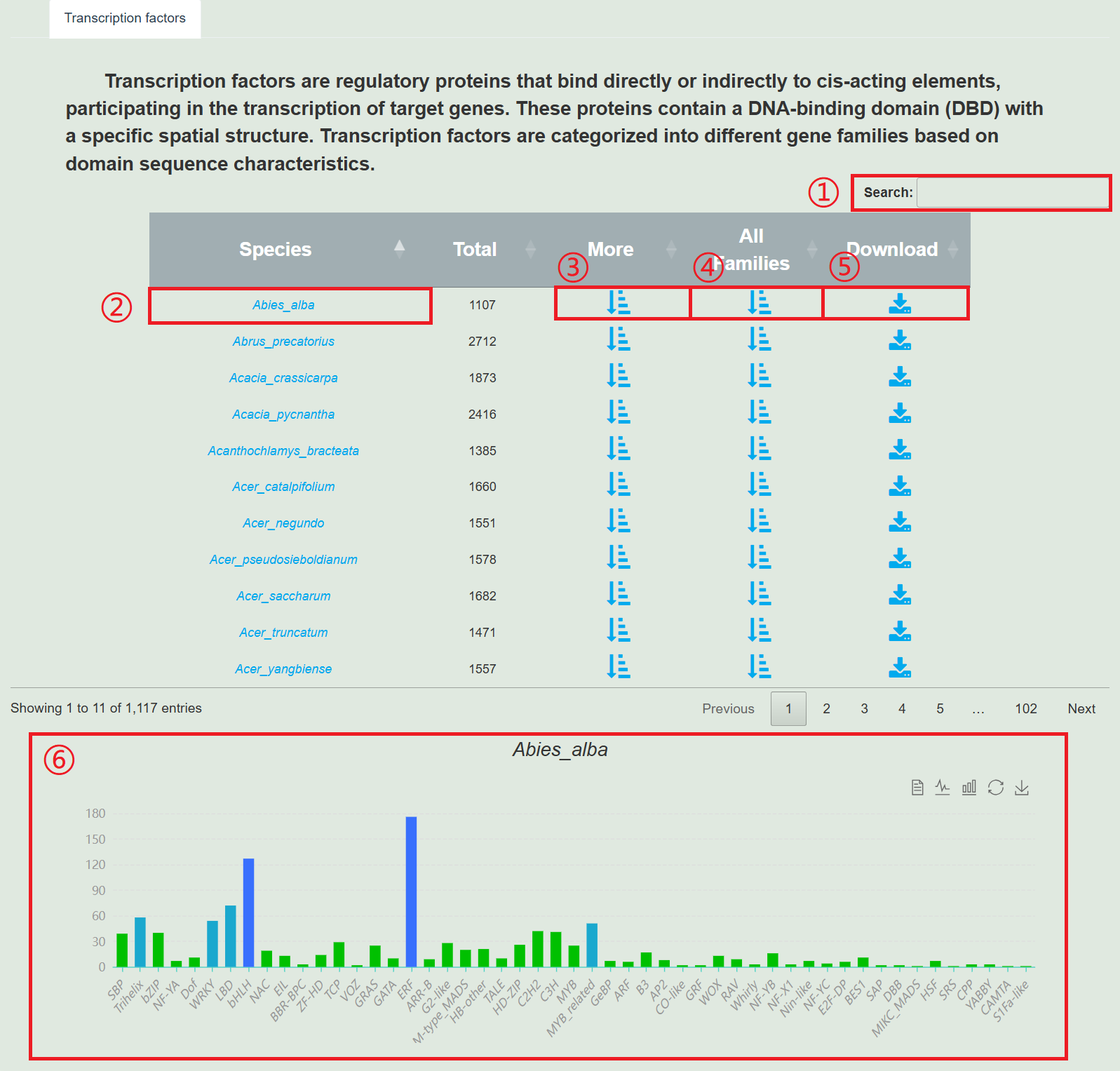
1.Search for species.
2.Clicking on the species name will redirect you to the species display page.
3.Click "More" to display all transcription factors of the species.
4.Click on 'All Family' to display the transcription factor family in the form of a bar chart.
5.Download all transcription factors of the species.
6.Column chart of species transcription factor family, click to display the transcription factors of this type.
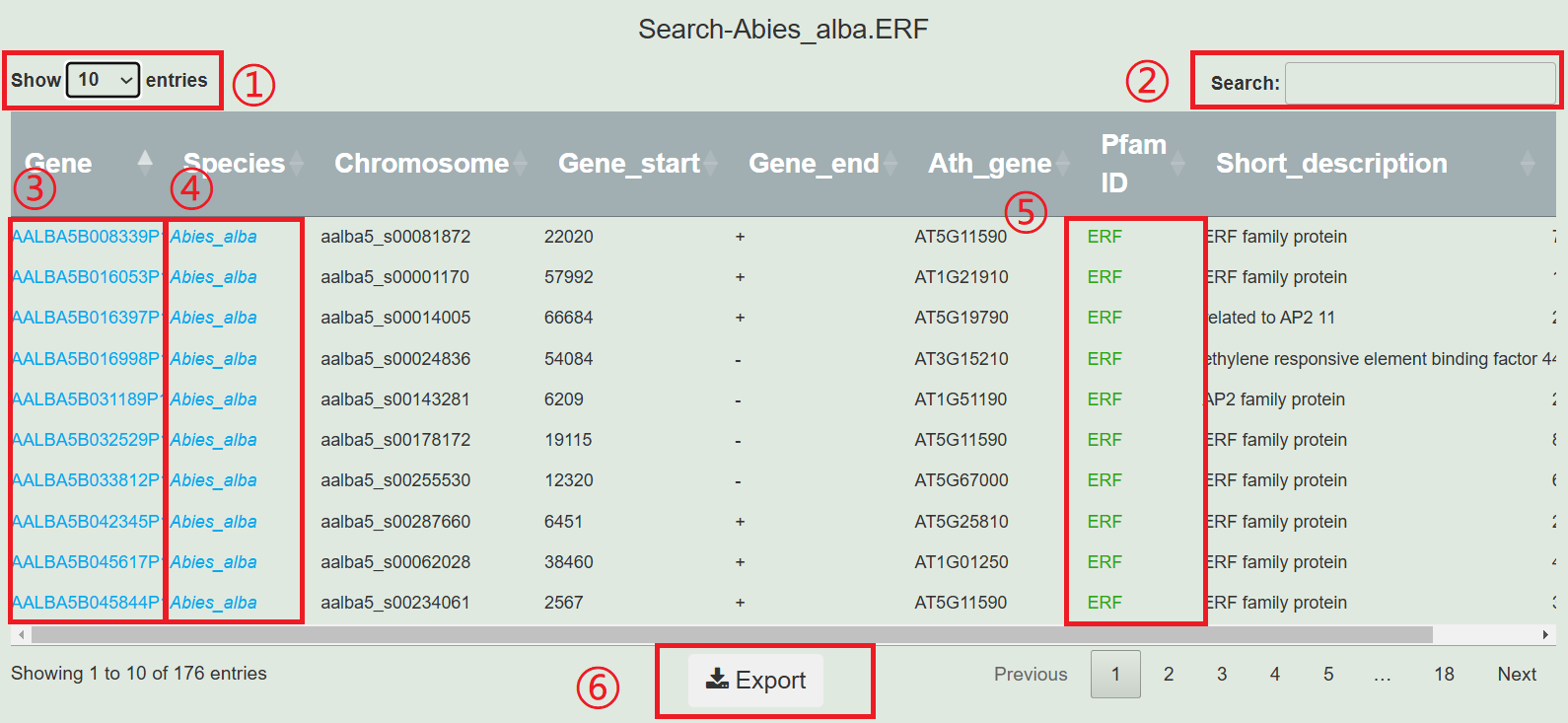
1.Select the number of displays.
2.Input genes, chromosomes, and description of interest for screening.
3.Click on the gene to jump to the gene display page.
4.Clicking on the species name will redirect you to the species display page.
5.Move the mouse over the description to view specific information.
6.Download all data from the table.
3. Annotations
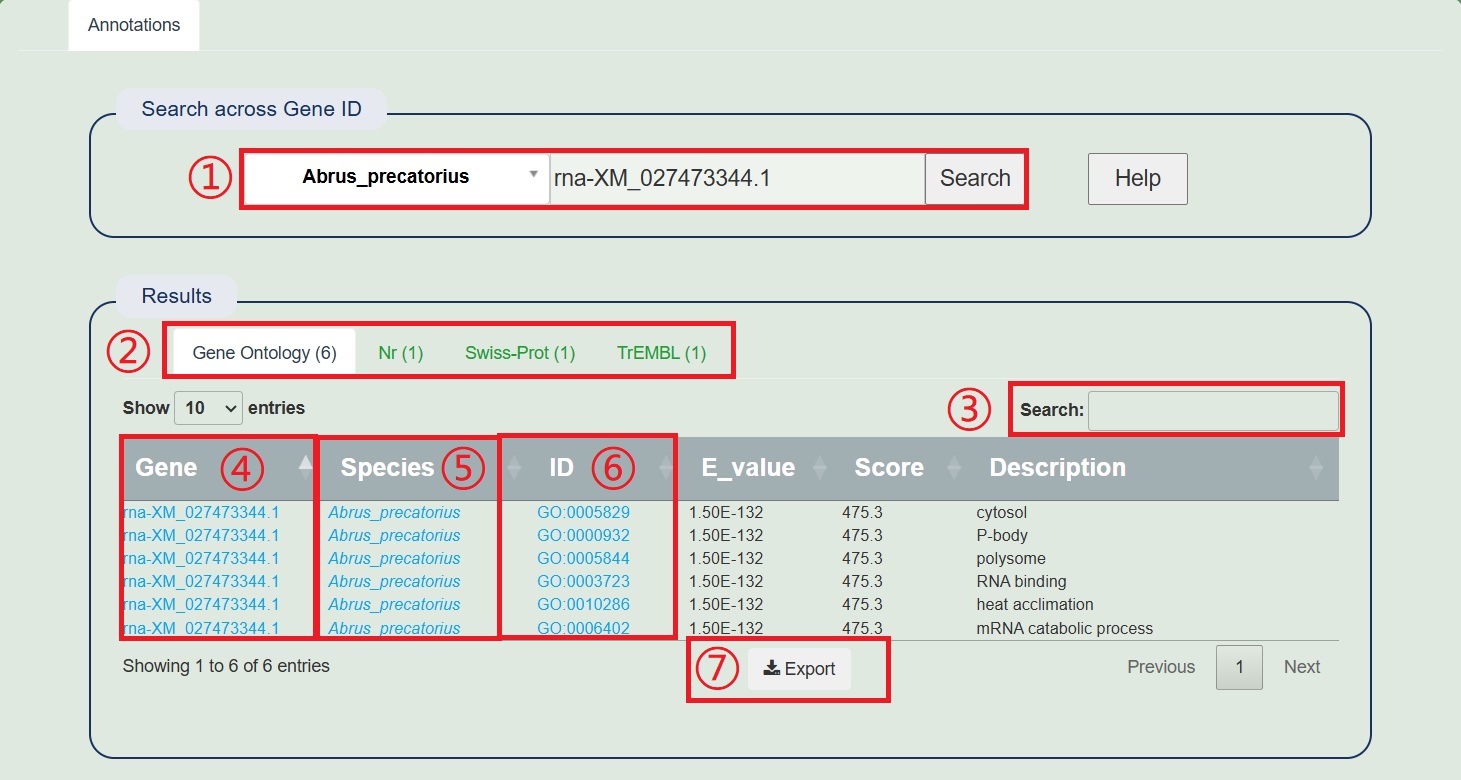
1.When entering a species, an example will pop up, and you can enter other genes again.
2.Switch between four types of annotations.
3.Input genes, and description of interest for screening.
4.Click on the gene to jump to the gene display page.
5.Clicking on the species name will redirect you to the species display page.
6.Click on the comment link to jump to the detailed page.
7.Download all data from the table.
1. Publication
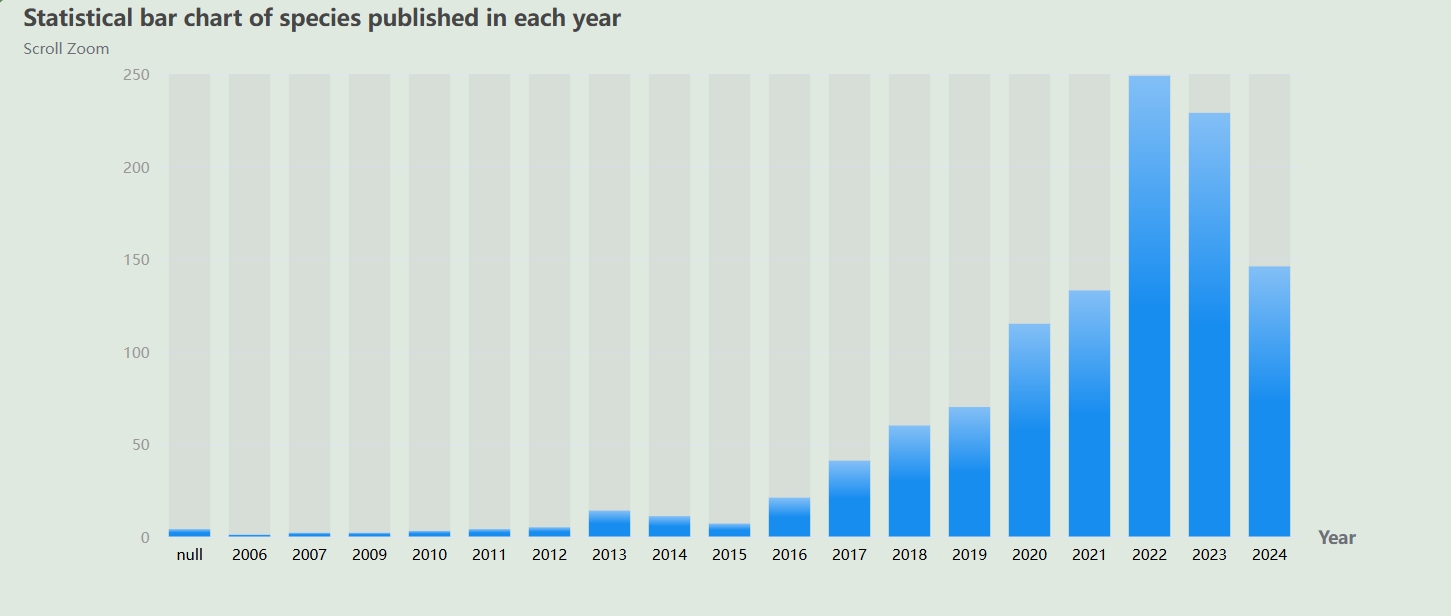
Statistical bar chart of species published in each year.Click to view specific literature
2. Database
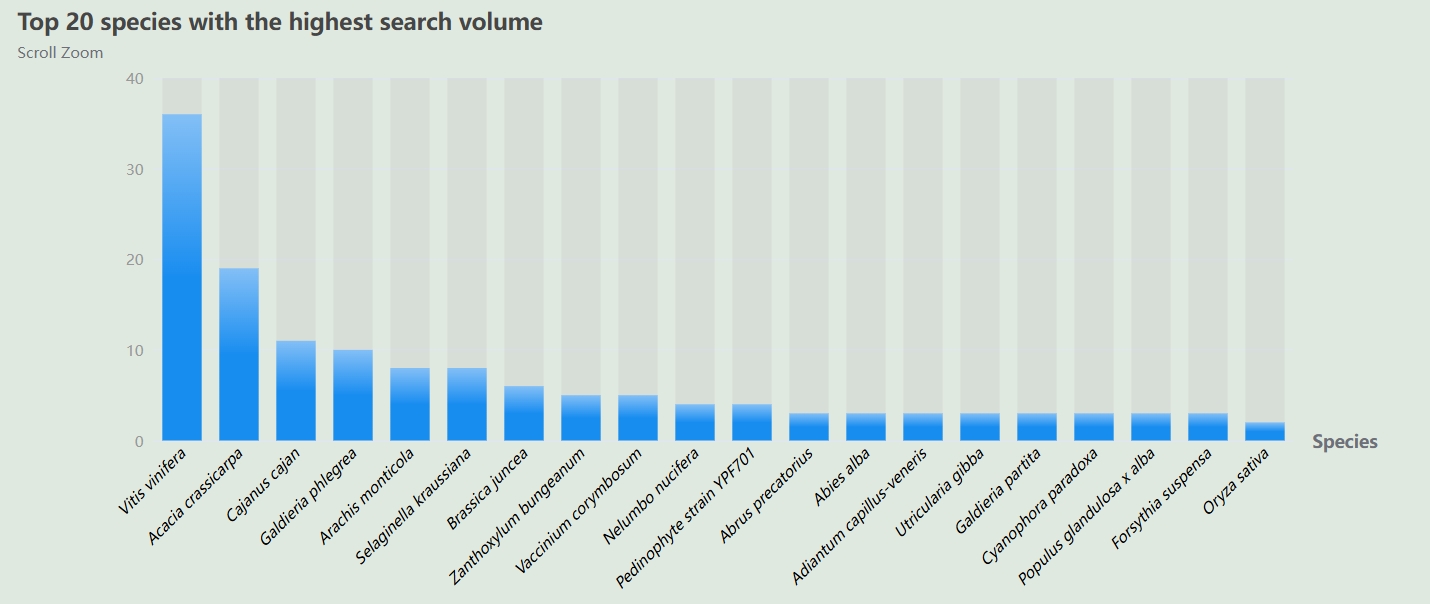
The database of data sources, click to view specific literature.
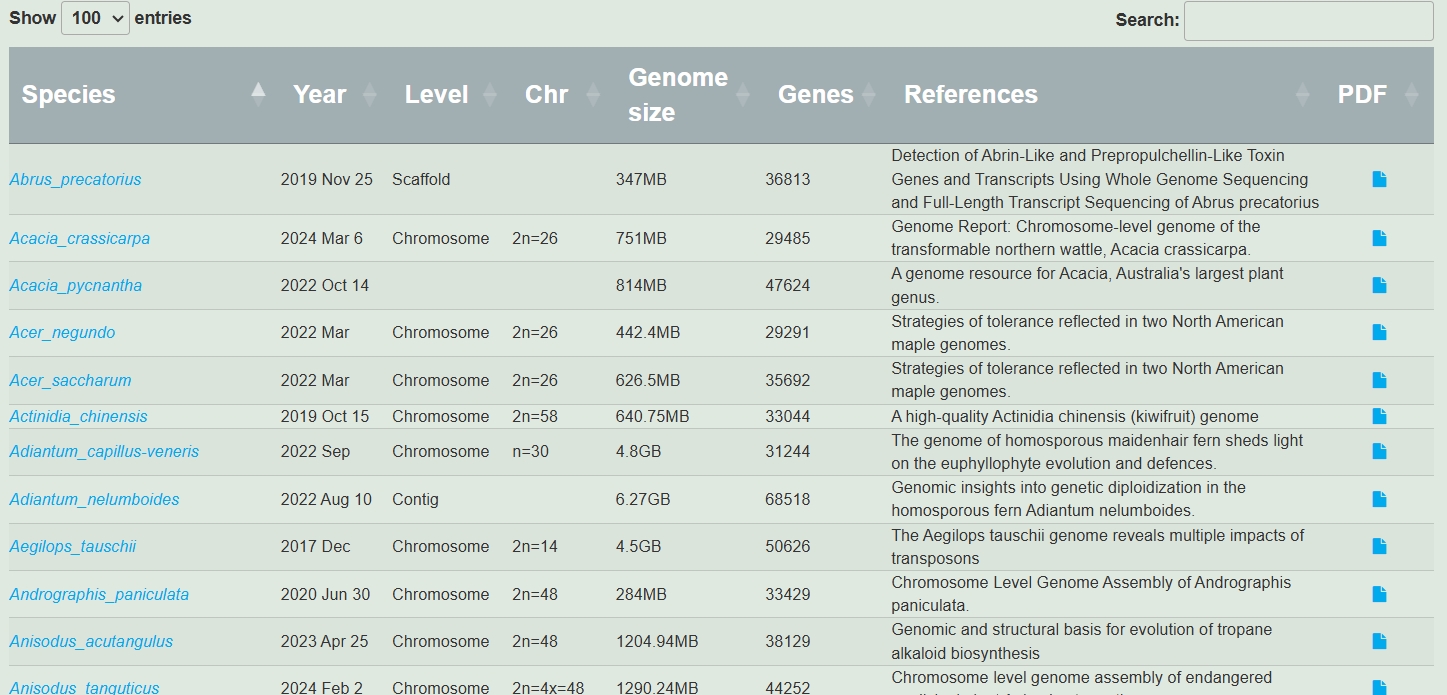
Specific literature and species information, including literature name, publication date, and genome size, etc.
1. Table list
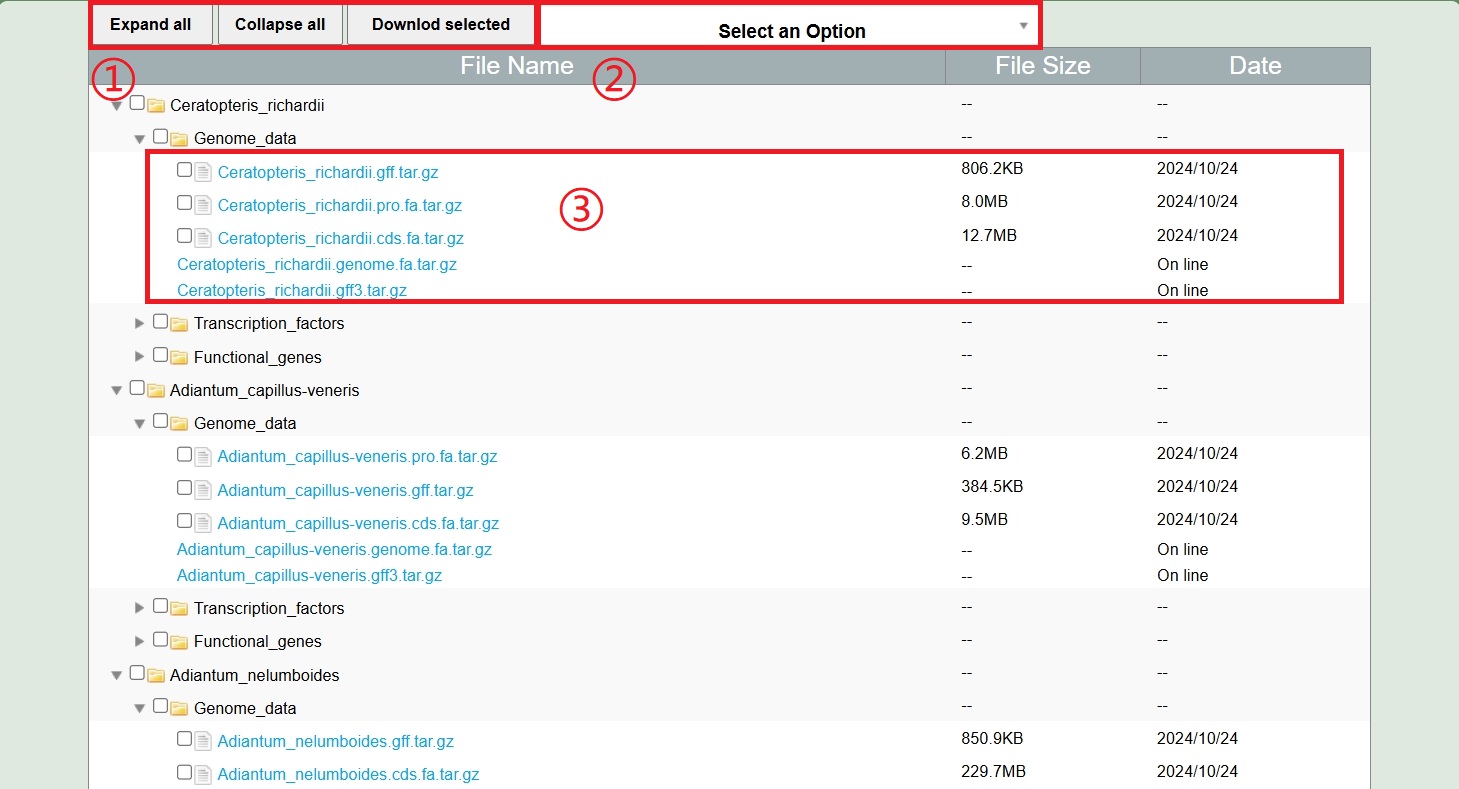
The download page contains five download files and two download folders of 1117 species: genome file, CDS file, Protein file, GFF3 file, GFF file (gene ID, chromosome number, start, stop, strand and gene ID), functional genes and transcription factors.
1.Expand all, shrink all, package and download selected files.
2.Search and select the species name, family, and genus to display the data table.
3.Display data information, click to download.
1.Expand all, shrink all, package and download selected files.
2.Search and select the species name, family, and genus to display the data table.
3.Display data information, click to download.
2. Download FTP
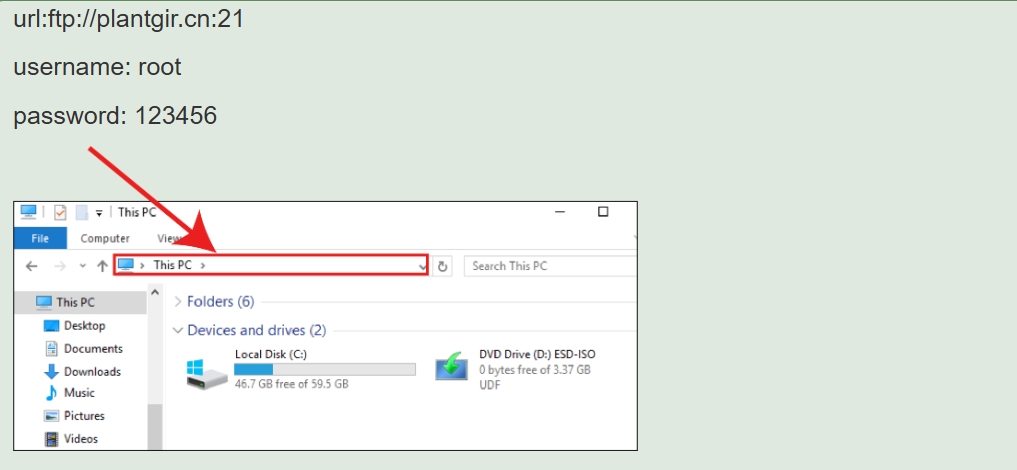
You can download files on your own computer through FTP (Online files are not included in the database and need to be downloaded using the first method).
3. Species tree
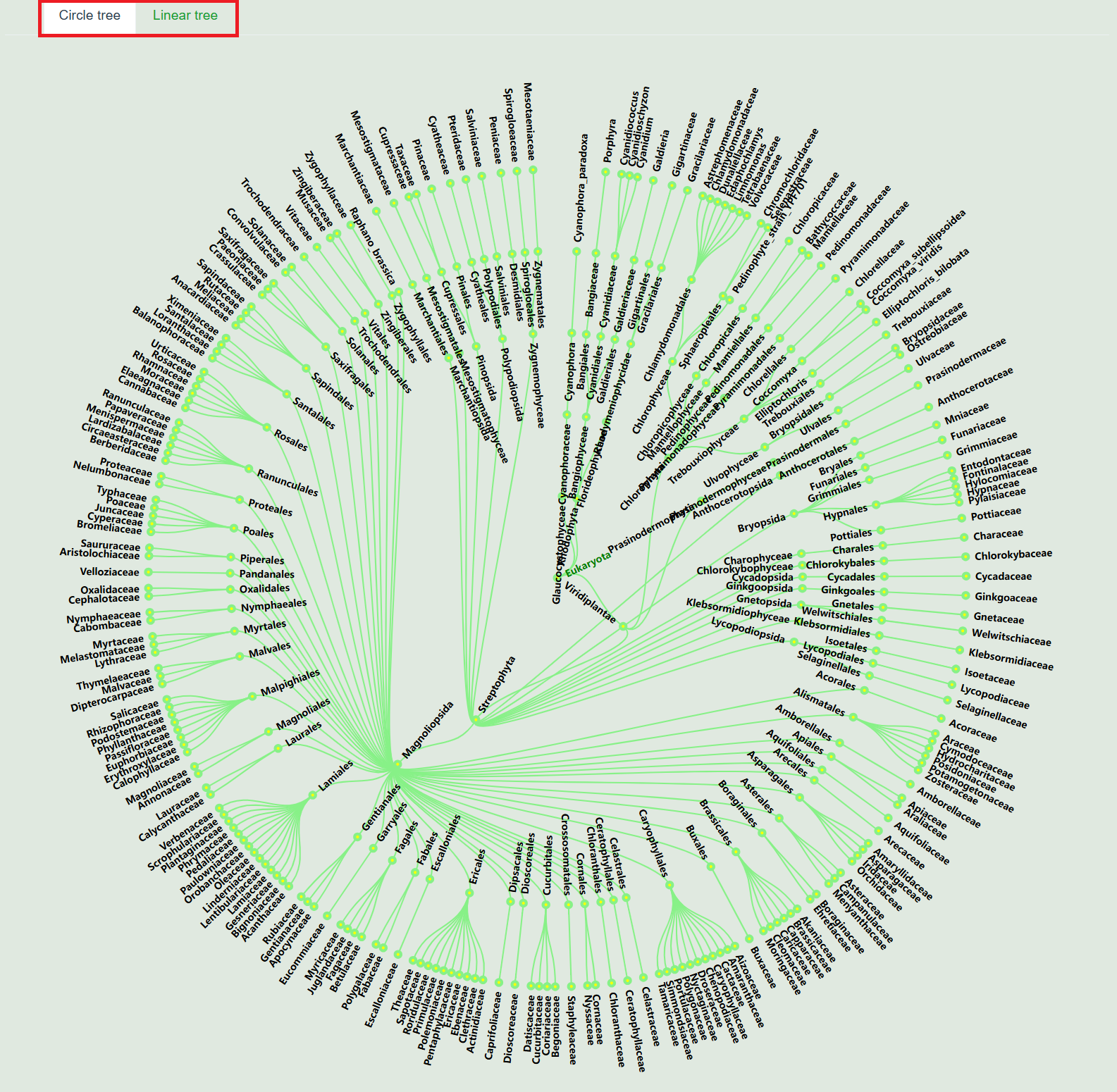
You can switch between circular and linear tree charts, and click on the species name to view species information and data.
1. Blast
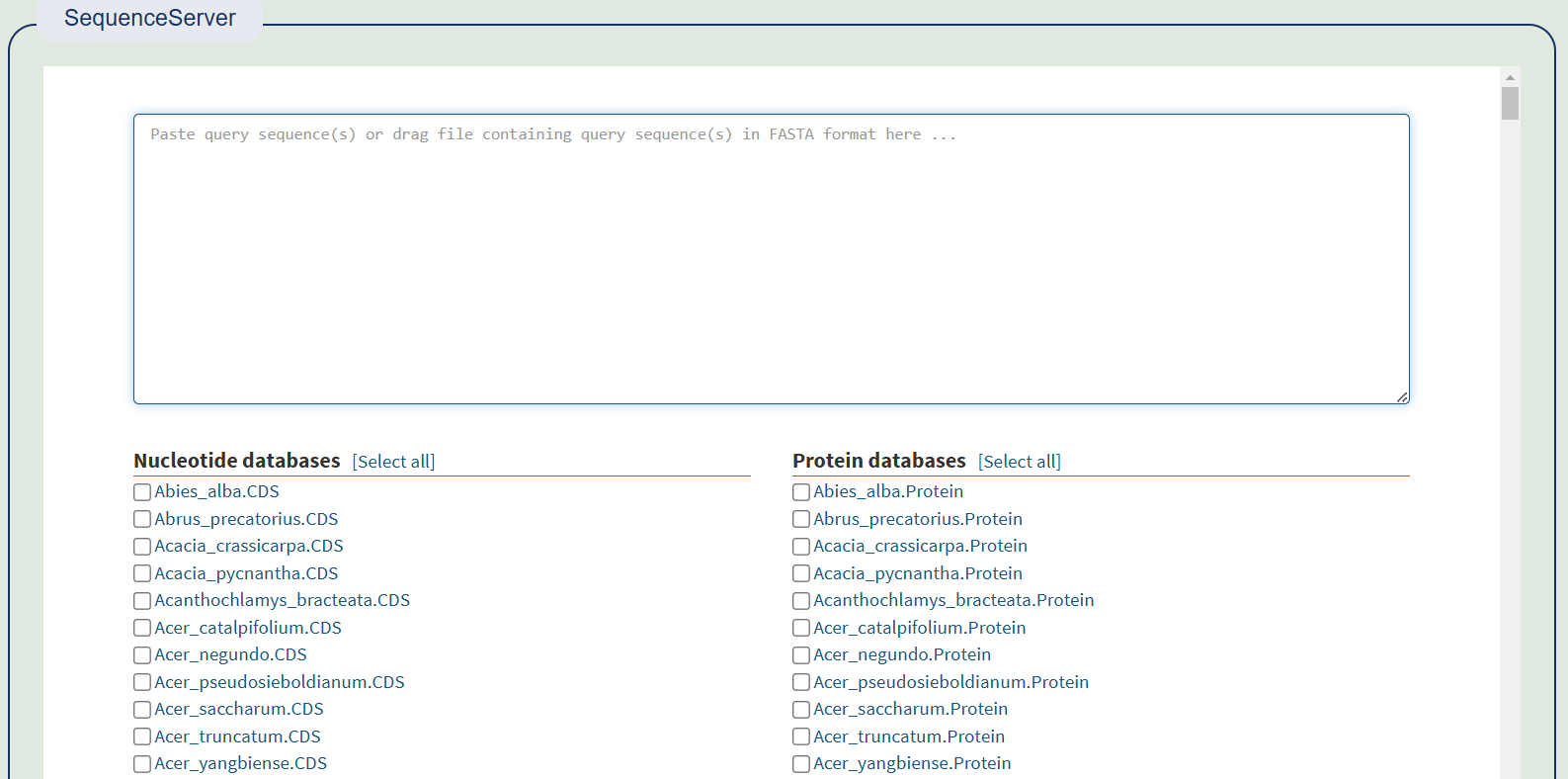
Blast uses the SequenceServer package and builds a library of CDS and proteins for all species, allowing users to freely choose and compare them.
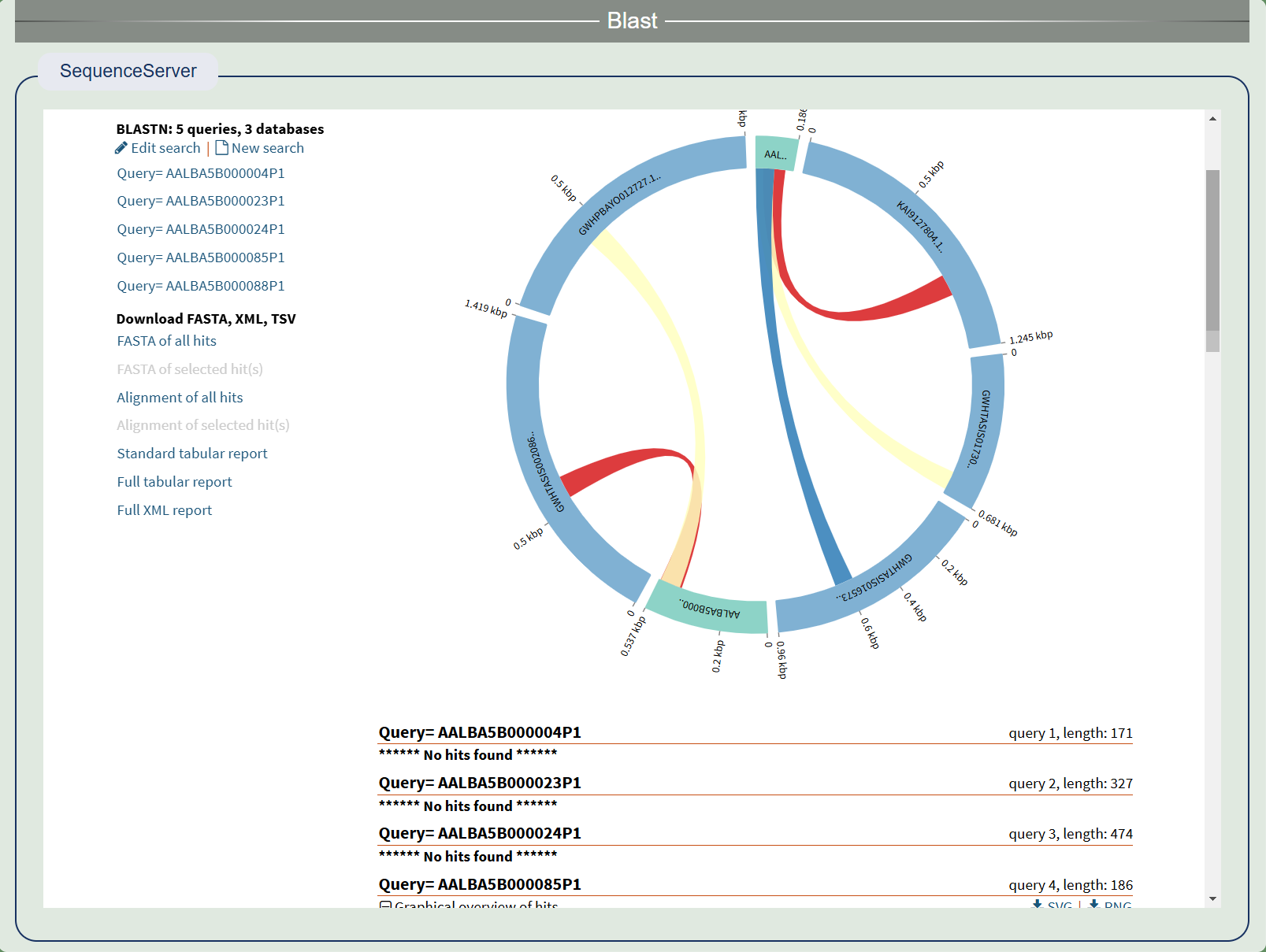
Comparison results of SequenceServer
2. Primer design
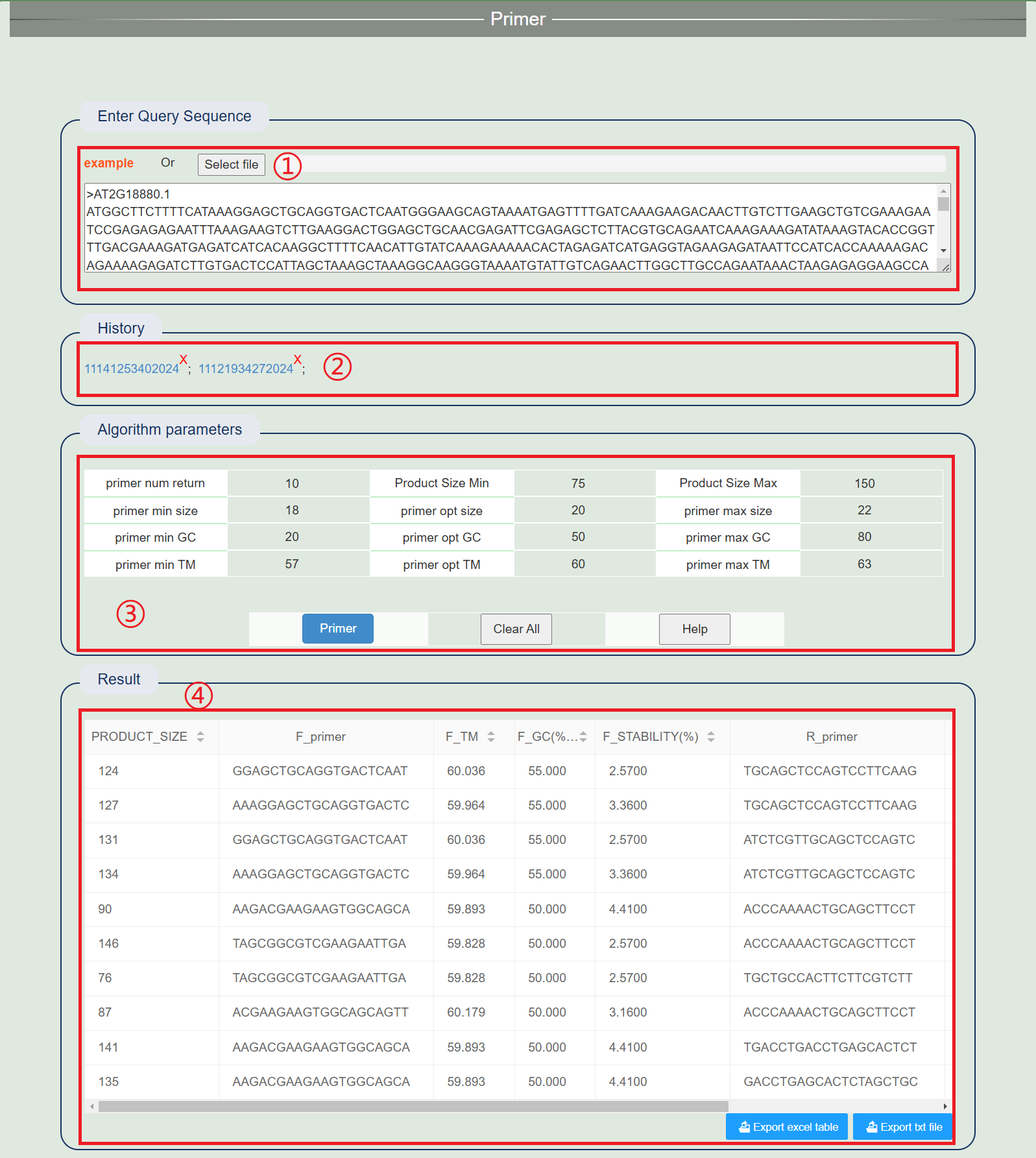
1. Click on the example or upload the file.
2. After successful operation, a history record will be generated. Clicking the link to view the results directly.
3. Select appropriate parameters.
4. Results include: Product size, F_primer, F_TM, F_FC, F_Stability and R_primer, R_TM, R_FC, R_Stability.
3. Hmmsearch
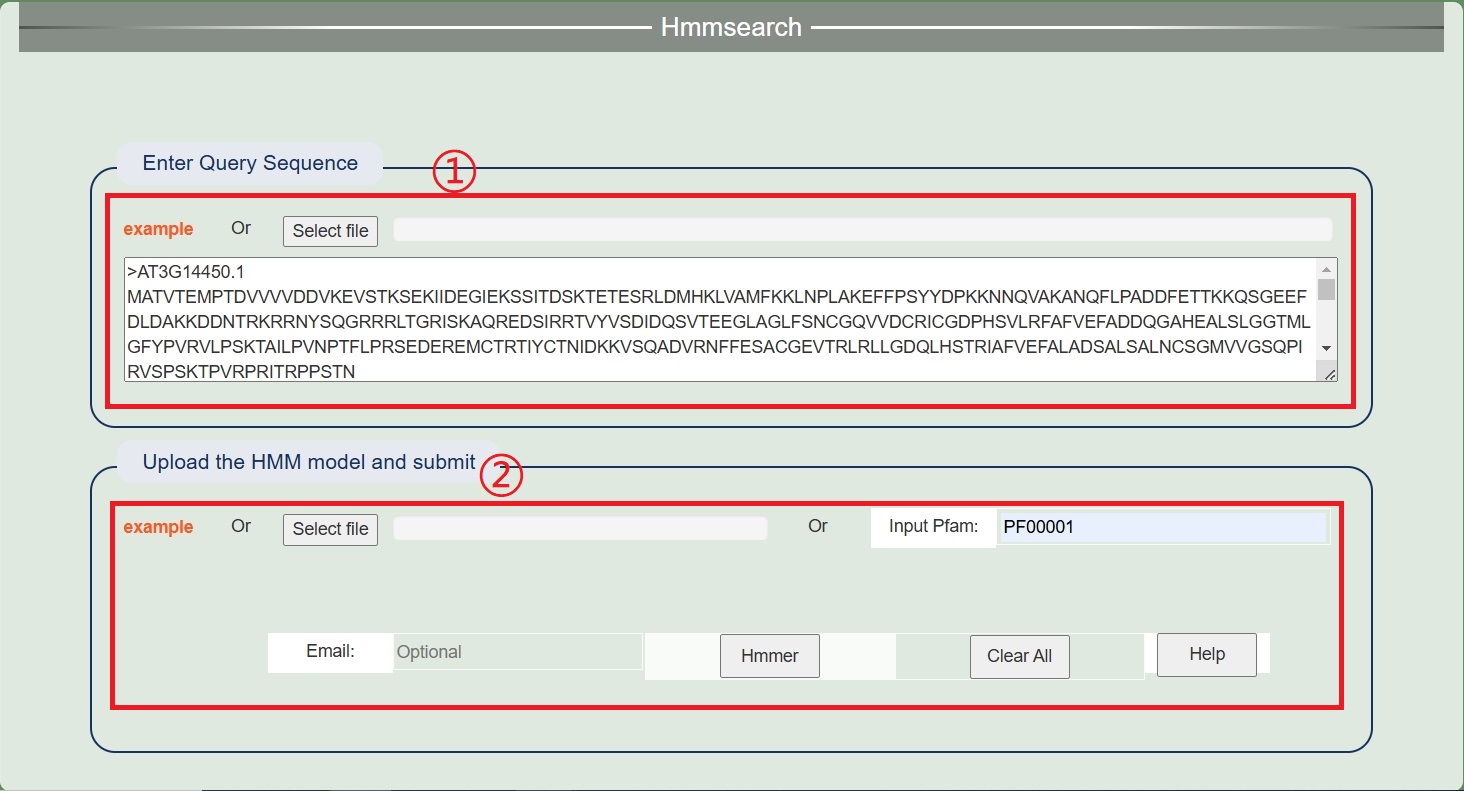
1.Enter an example file or submit a sequence file.
2.Select online models or submit your own models. Results will be downloaded directly as files.
4. Sequence alignment
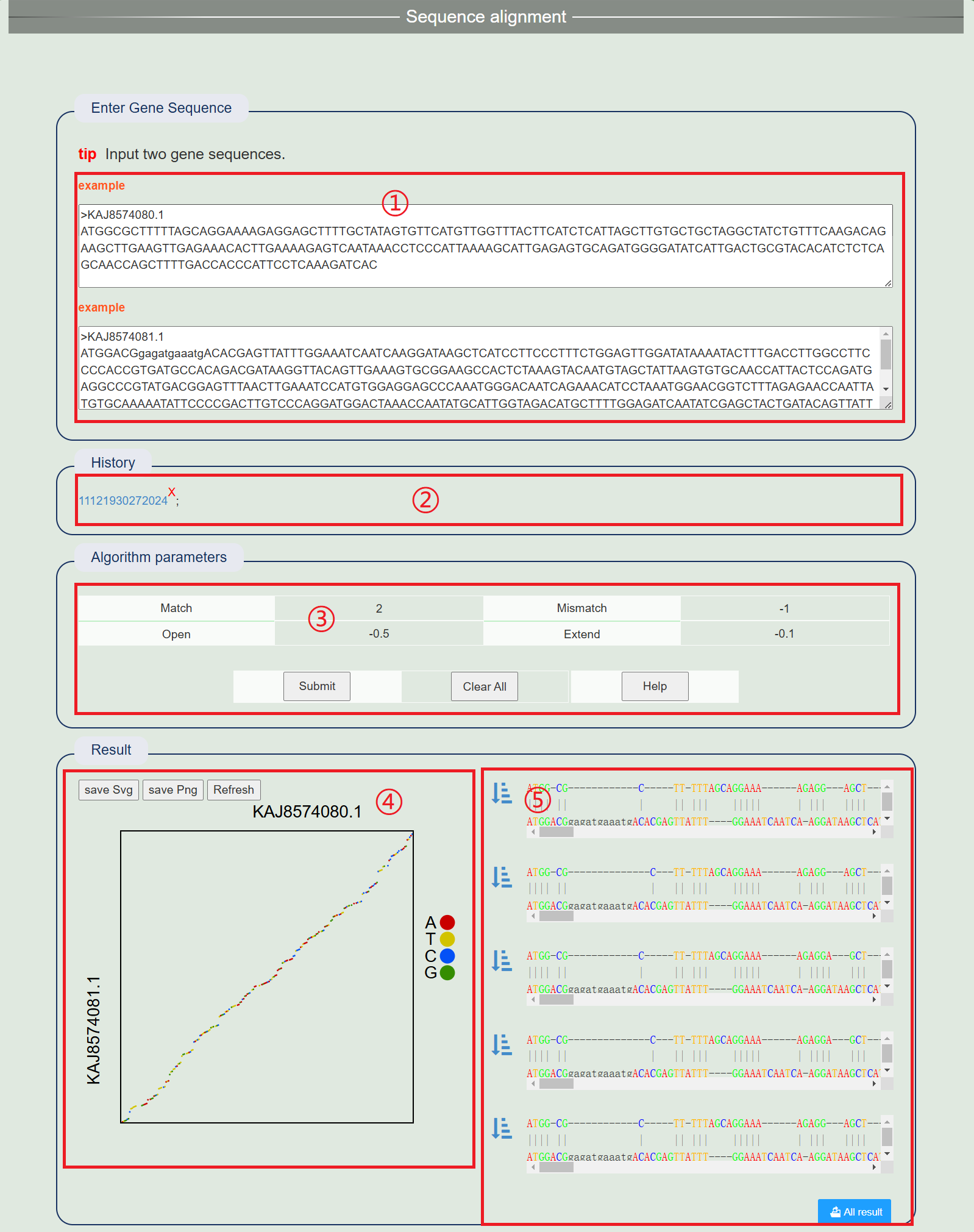
1. Enter the CDS sequence to be compared.
2. After successful operation, a history record will be generated. Click the link to view the results directly.
3. Adjust comparison parameters.
4. View the dot map at the level of the first base that is best compared in the results, with different colors representing four different bases. Click on the far point on the right to display or cancel the display of this base.
5. View the first five comparison results. Click the left eye icon to view the dot map made of specific bases on the left.
1. Functional annotation of genes
Gene annotations of 59 vegetable species were conducted using five protein databases, including Swiss-Prot and TrEMBL of UniProt knowledgebase( https://www.uniprot.org ),Pfam(v34.0)( http://pfam.xfam.org . Gene Ontology (go, http://geneontology.org )and non-redundant protein sequence database (NR, https://www.ncbi.nlm.nih.gov ). The annotation information of all genes was obtained by blastp with 59 vegetables with Arabidopsis thaliana.
2.Identification of TFs and functional genes
The 63 TF gene families were detected, using the Pfam database, from the protein sequences of 59 vegetable species (E-value < 1e-5). The 295 flowering genes of Arabidopsis were collected from previous studies and the FLOR-ID database. The 73 glucosinolate genes of Arabidopsis were collected from the previous reports, and the 41 anthocyanin genes and 151 auxin genes of Arabidopsis were obtained from the BRAD website. Homologous flowering, glucosinolate, anthocyanin, and auxin genes in other vegetable species were identified by Blastp (E-value < 1e-5; Identity >60%; Score >150) with manual checking.
Telephone: 0315-8805607
Mail: songxiaoming116@163.com
Location: 21 Bohai Road, Caofeidian Xincheng, Tangshan 063210, Hebei, China
Working Hours:Mon - Fri: 8:00 am to 10:30 pm
Mail: songxiaoming116@163.com
Location: 21 Bohai Road, Caofeidian Xincheng, Tangshan 063210, Hebei, China
Working Hours:Mon - Fri: 8:00 am to 10:30 pm
Some pictures and data of this website come from the Internet and other public database platforms. Copyright belongs to the original author. If there is any infringement, please contact Song Xiaoming (songxiaoming116@163.com) in time to delete it. In addition, all data in this database is free for all scientists to use. For commercial use, please contact us for authorization.